Development of reagents for liquid biopsy-based diagnostic stratification and targeted treatment of breast tumors
The project was supported by Javna Agencija za Raziskovalno dejavnost Republike Slovenije (grant ARRS/J4-9322). Composition of the project team: Ario de Marco (SICRIS ID: 32943), Elisa Mazzega (SICRIS ID: 50086), Sandra Oloketuyi (SICRIS ID: 54863), Polona Bedina (SICRIS ID: 16104)
Abstract
The capacity to identify biologically distinct tumor sub-types according to the expression of specific biomarkers has critically improved the therapy efficiency and minimized the toxicity caused by applying ineffective therapies. It is expected that the progressive integration of the present knowledge with further biomarkers will increase the chance to design optimal targeted therapeutic strategies for any biological profile. Particularly critic would be the implementation of systematic diagnostic screenings to identify early events and prevent the disease development into advanced primary and metastatic cancers for which the present therapies are still highly ineffective. This approach would provide the necessary information for optimized precision therapies and significantly decrease the social and economic burden of cancer. The progress pace will depend on the success in the identification of new biomarkers suitable for more precise tumor stratification and the successive development of reliable binders for selective recognition of tumor biomarkers on cells and in biological fluids. Affordable large-scale tests to detect early stage tumor in (risk) population will be however feasible if the diagnostic methodology will be not only reliable and sensitive, but also rapid, non-invasive, and inexpensive. In this perspective, Liquid Biopsy based on the evaluation of exosome biomarkers has the capacity to recapitulate the patient diagnostic status because this extracellular vesicle (EVs) class proved being involved in metastasis, and they are easily accessible in any body fluid. At the present, the technological limiting factor is the exosome purification protocol and our proposal aims at overcoming this bottleneck by developing antibodies for exosome immunopurification. Immunocapture is the method which provides the most pure exosome fractions and the only one available for separating extracellular vesicle sub-populations, such as tumor-specific exosomes from the “background” (physiological) exosomes present in biological fluids. Antibodies selective for exosome sub-populations will enable their fractionation and to evaluate separately their molecular content to identify the diagnostic relevant biomarkers. The exclusive recovery of tumor-related macromolecules will also contribute understanding the biological mechanisms by which exosomes participate into tumor progression, long-distance cell-to-cell regulation, and tumor microenvironment modifications. We have already demonstrated that recombinant antibody technology is suitable for the fast recovery of reliable anti-exosome binders and now propose the implementation of a platform for the production of an array of single-domain antibodies (also called nanobodies or VHHs) engineered for tumor diagnostic applications.
Results
Nanobodies specific for antigens expressed on the surface of triple-negative breast cancer cells and extracellular vesicles (EVs) were isolated challenging a pre-immune library first with control samples and then the unbound fraction with the target samples. The next steps were:
i) the optimization of the nanobody display; ii) the engineering of report moieties; iii) the maturation of the initial binders into second-generation reagents; iv) the development of a biosensor for biomarker quantification; v) the upscale of EV purification
- i) the diagnostic exploitation of nanobodies can consider solutions that differ in terms of production effort and costs. The cheapest and fastest solution is their display on bacteria to obtain “living fluo-immunoreagents” (Figure 1)

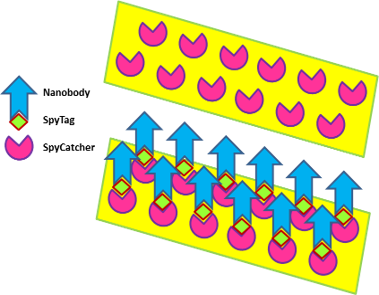
- ii) we adopted different strategies for nanobody labeling, starting with conventional ones (Avitag, SNAP, sortase, free cysteine,…) and finally chose two approaches, either the direct fusion with fluorescent proteins or the SpyTag/SpyCatcher system (Figure 2)
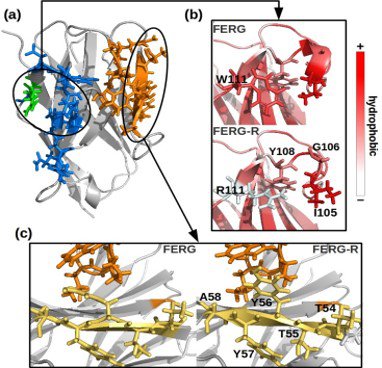
- iii) nanobodies recovered by panning can be considered hits which need to be optimized into lead reagents. Random mutagenesis has been used in the past to improve specific characteristics of ligands but is there a rational maturation process? The short sequence of nanobodies is particularly suitable for in silico modeling and succeeded in deciphering some aspects regulating the nanobody biophysical features (Figure 3)
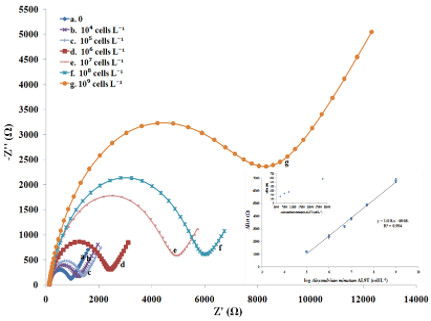
- iv) Exact quantification of even minimal amounts of targets in biological samples is a prerequisite for reliable diagnostics and immunocapture provide the necessary selectivity. Based on these two principle, we built a biosensor with a limit of detection of 3 cells/mL and a dynamic range spanning over 5 orders of magnitude (Figure 4)
- v) the purification upscale of specific classes of EVs is still an open problem. We gave our contribution by designing a conceptually new chromatographic method based on nanobody-activated monoliths (Figure 5)
Publications
- Oloketuyi S, Dilkaute C, Mazzega E, Jose J, de Marco A (2019) Purification-independent immunoreagents obtained by displaying nanobodies on bacteria surface. Appl Microbiol Biotechnol, 103:4443-4453
- Soler M, Medagli B, Semrau M, Storici P, Bajc G, de Marco A, Laio A, Fortuna S (2019) A consensus protocol for the in silico optimisation of antibody fragments. Chem Comm, 55: 14043-14046
- Veggiani G, Giabbai B, Semrau MS, Medagli B, Riccio V, Bajc G, Storici P, de Marco A (2020) Comparative analysis of fusion tags used to functionalize recombinant antibodies. Prot Expr Purif, 166:105505
- Bernardinelli G, Oloketuyi S, Werner SF, Mazzega E, Högberg B, de Marco A (2020) A compact nanobody-DNAzyme conjugate enables antigen detection and signal amplification. N Biotechnol, 56:1-8
- Oloketuyi S, Mazzega E, Zavašnik J, Pungjunum K, Kalcher K, de Marco A, Mehmeti E (2020) Electrochemical immunosensor functionalized with nanobodies for the detection of the toxic microalgae Alexandrium minutum using glassy carbon electrode modified with gold nanoparticles. Biosensors Bioelectronics, 54:112052
- de Marco A (2020) Recombinant expression of nanobodies and nanobody-derived immunoreagents. Prot Expr Purif 172:105645
- Ubbiali D, Orlando M, Kovačič M, Iacobucci C, Semrau MS, Bajc G, Fortuna S, Ilc G, Medagli B, Oloketuyi S, Storici P, Sinz A, Grandori R, de Marco A (2021) An anti-HER2 nanobody binds to its antigen HER2 via two independent paratopes. Int J Biol Macromol,182:502-511
- Soler MA, Medagli B, Wang J, Oloketuyi S, Bajc G, Huang H, Fortuna S, de Marco A (2021) Effect of humanizing mutations on the stability of the llama single-domain variable region. Biomolecules 11:163
Project period: July 2018 to June 2021

